14. Adding Rivers#
If you want to include rivers in your simulation domain, there are several variables to define as:
the number of rivers: Nsrc
the position of the rivers on the model grid: Isrc and Jsrc
the zonal or meridional axis of the river flow: Dsrc
if flow (and concentration) is constant, the flow rate of the river (in m3/s): Qbar (positive or negative)
if flow (and concentration) is variable, and read from a netCDF file, the direction of the flow qbardir:
1 for west-east / south-north
-1 for east-west / north-south
the type of tracer advected by the river: Lsrc
the value/concentration: Tsrc
14.1. Constant flow and concentration#
For this you need to define the cpp-keys in cppdefs.h
#define PSOURCE
And re-compile.
Then in the croco.in file
psource: Nsrc Isrc Jsrc Dsrc Qbar [m3/s] Lsrc Tsrc
2
3 54 1 200. T T 20. 15.
3 40 0 200. T T 20. 15.
where Nsrc=2 is the number of rivers processed, then each line describes a river. Let’s describe the parameter for river #1:
Isrc=3, Jsrc=54 are the i, j indices where the river is positioned
Dsrc=1 indicates the orientation (here meridional => along V direction)
200 is the runoff flow value in m3/s, oriented to the east
T T are true/false indications for reading or not the following variables (here temperature and salinity)
20 and 15 are respectively the temperature and salinity of the river. You can edit these parameters.
Warning
The sources points must be placed on U or V points on the C-grid and not on rho-points
You can then run the model:
qsub job_croco_mpi.pbs
14.2. Variable flow read in a netCDF file and constant concentration#
Instead of using a constant flow, you can use variable flow. For that you need read it from a netcdf file. First define the dedicated cpp-key in cppdefs.h
#define PSOURCE_NCFILE
And re-compile the model.
Then you also need to prepare the netcdf river runoff input file.
For that, you can use in CROCO_TOOLS make_runoff (Rivers/make_runoff.m) which detect the main rivers located in your domain (from RUNOFF_DAI runoff climatology).
Note
RUNOFF_DAI is a global monthly runoff climatology containing the 925 first rivers over the world, from Dai and Trenberth, 2000
After asking you some specifications for each detected river in your domain, for the selected rivers:
It will compute the right location on the croco_grid regarding the direction and orientation you defined
It will create the river forcing netCDF file croco_runoff.nc containing the various river flow time series.
To do so, in CROCO_TOOLS, edit make_runoff.m and define the following flags:
%% Choose the monthly runoff forcing time and cycle in days
clim_run=1
% - times and cycles for runoff conditions:
% - clim_run = 1 % climato forcing experiments with climato calendar
% qbar_time=[15:30:365];
% qbar_cycle=360;
%
% - clim_run = 0 % interanual forcing experiments with real calendar
% qbar_time=[15.2188:30.4375:350.0313];
% qbar_cycle=365.25;
psource_ncfile_ts=0;
% - psource_ncfile_ts = 0 => Constant analytical runoff tracers concentration no processing
% It reads analytical values in croco.in
% or use default value defined in
% analytical.F
For the BENGUELA test case, you will have 2 rivers detected, Orange and Doring. We recommend to define them as zonal (0) and oriented from east to west (-1). It will give you the lines to enter in the croco.in file in the psource_ncfile section.
psource_ncfile: Nsrc Isrc Jsrc Dsrc qbardir Lsrc Tsrc runoff file name
CROCO_FILES/croco_runoff.nc
2
25 34 0 -1 30*T 20 15
31 19 0 -1 30*T 20 15
where Nsrc=2 is the number of rivers, then each line describe a river. Let’s describe the parameter for the river #1
Isr=25, Jsrc=34 are the i, j indices where the river is positioned
Dsrc=0 indicates the orientation (here zonal)
qsbardir= -1 indicates the direction (here towards the west)
Lstrc=30*T are true/false flags for reading or not the following variables (here temperature and salinity)
Tsrc=20 15 are respectively the temperature and salinity of the river.
You can edit these parameters.
Temperature and salinity can also be variable and read from a netCDF file, it is described in the next section.
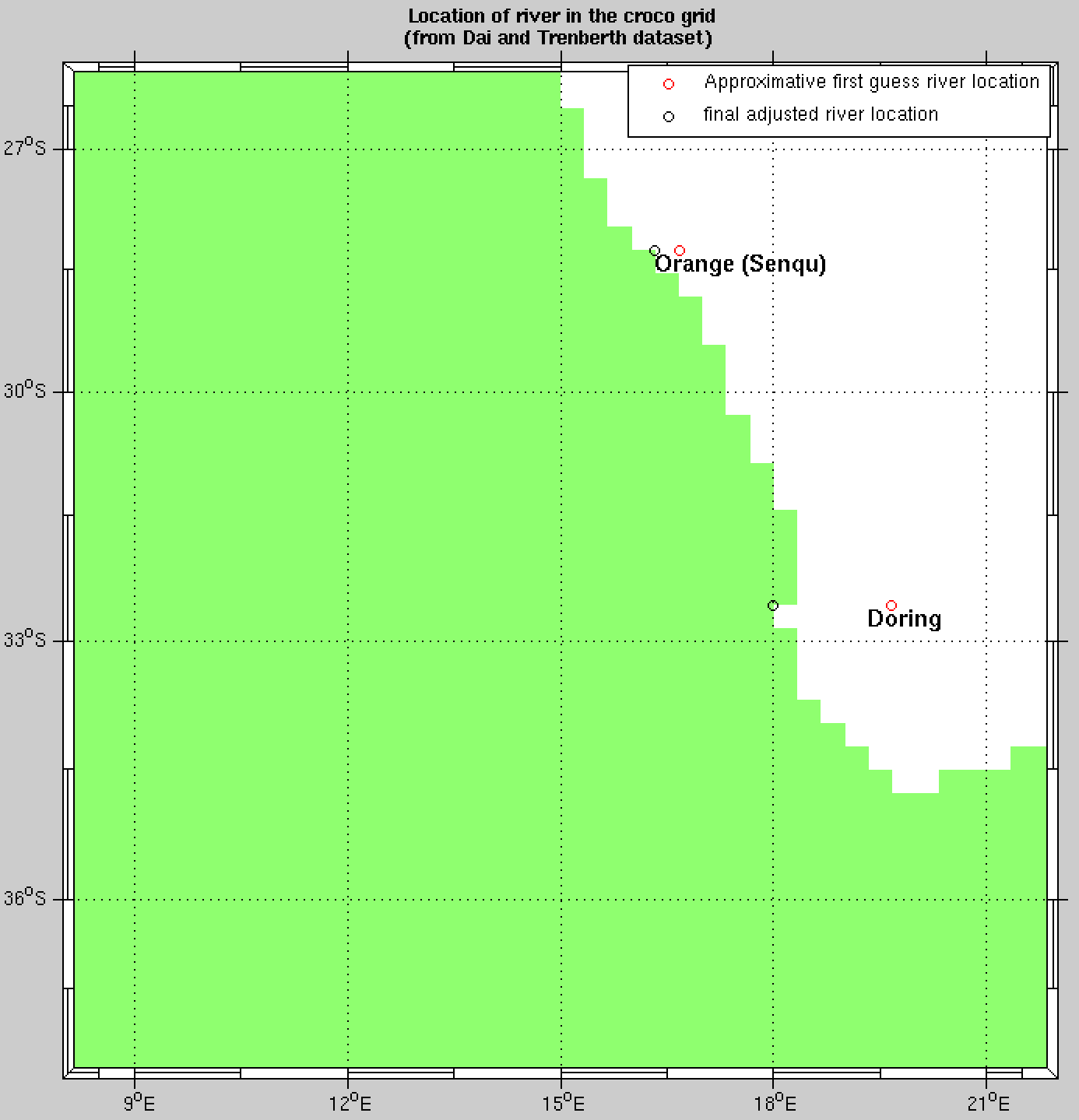
First and final guess rivers positions#
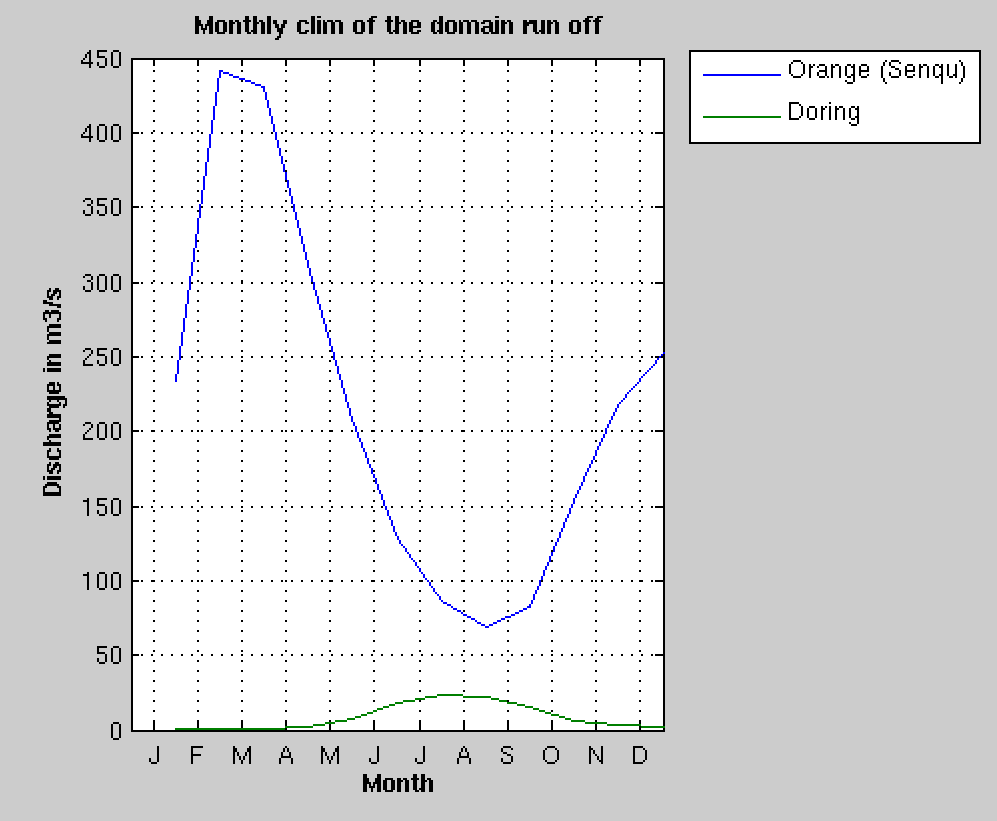
Rivers flow seasonal cycle#
14.3. Variable flow and variable concentration from a netCDF file#
To run CROCO with a variable concentration of river tracers, you need to define the following cpp-key in cppdefs.h
#define PSOURCE_NCFILE_TS
You also need to prepare your netcdf input file. Using the CROCO_TOOLS: edit make_runoff.m and change the following flags:
psource_ncfile_ts=1;
if psource_ncfile_ts
psource_ncfile_ts_auto=1 ;
psource_ncfile_ts_manual=0;
end
% - pource_ncfile_ts = 1 => Variable runoff tracers
% concentration processing is activated.
%
% In this case, either choose:
% - psource_ts_auto : auto definition
% using the nearest point in the climatology
% file croco_clm.nc to fill the tracer
% concentration time serie in croco_runoff.nc
%
% - psource_ts_manual : manually definition the
% variable tracer concentration to fill
% the tracer concentration time serie in
% croco_runoff.nc
After asking you some specifications of each detected river in your domain, for the selected rivers, in addition to river flow as in previous section, it will also put the tracers concentration (temp,salt, no3, et …) time series into the river forcing netCDF file croco_runoff.nc
psource_ncfile: Nsrc Isrc Jsrc Dsrc qbardir Lsrc Tsrc runoff file name
CROCO_FILES/croco_runoff.nc
2
25 34 0 -1 30*T 16.0387 25.0368
30 19 0 -1 30*T 16.1390 25.1136
You also can edit these parameters.
Warning
The Tsrc value reported in croco.in are the annual-mean tracer values, the are just for information. The real tracer concentration (Tsrc) are read in the runoff netCDF file created.
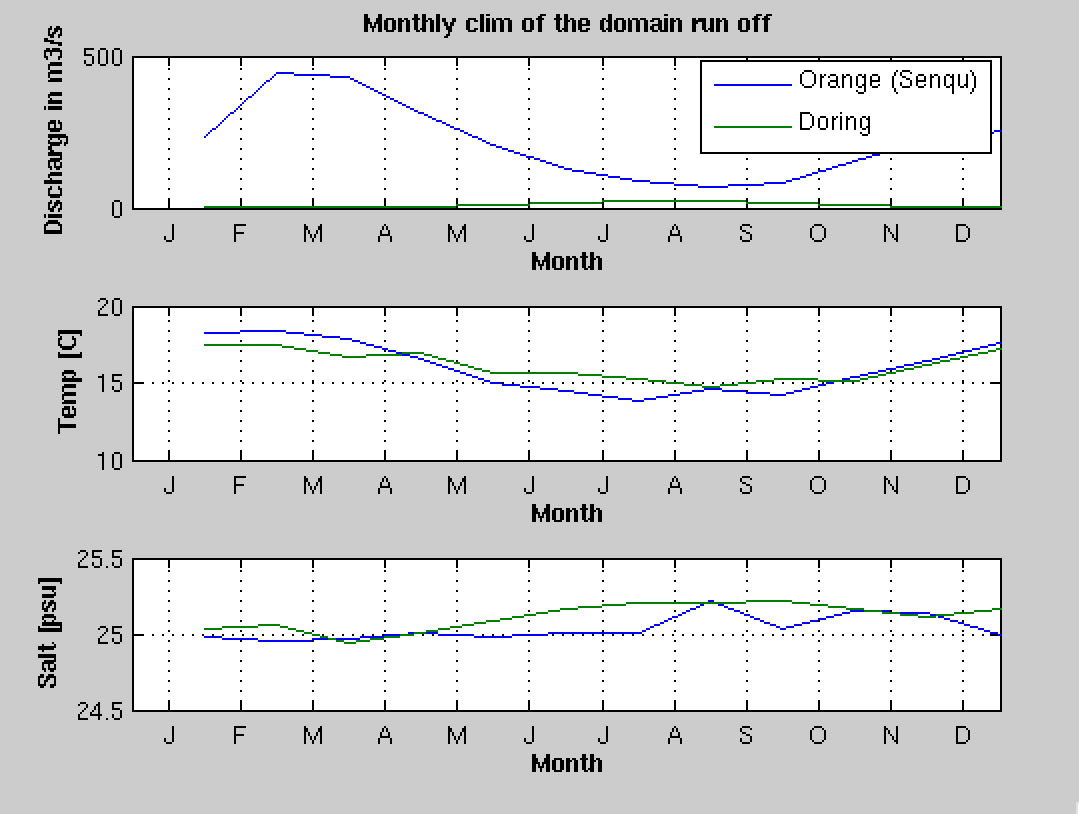
Rivers tracer concentration seasonal cycle#
14.4. Using a nest#
The above procedure can be applied to a nested grid. For this, edit make_runoff and change the gridlevel variable to the adhoc grid level.
%Choose the grid level into which you ant to set up the runoffs
gridlevel=1
if ( gridlevel == 0 )
% -> Parent / zoom #O
grdname = [CROCO_files_dir,'croco_grd.nc'];
rivname = [CROCO_files_dir,'croco_runoff.nc']
clmname = [CROCO_files_dir,'croco_clm.nc']; % <- climato file for runoff
else
% -> Child / zoom #XX
grdname = [CROCO_files_dir,'croco_grd.nc.',num2str(gridlevel)];
rivname = [CROCO_files_dir,'croco_runoff.nc.',num2str(gridlevel)];
clmname = [CROCO_files_dir,'croco_clm.nc.',num2str(gridlevel)]; % <- climato file for runoff
end
and run make_runoff again to generate
croco_runoff.nc.1
Note
The runoff has a default vertical profile defined in CROCO as an exponential vertical distribution of velocity. It is in analytical.F, subroutine ana_psource if you need to change it.
